AMPHIPOLS: Trapping membrane proteins with A8-35
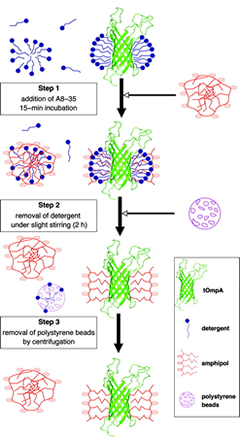
Although some exceptions have been encountered (reviewed in ref. 1), A8-35 as a rule will not extract by itself a MP from its native environment. The standard procedure is to solubilize the target protein in a classical detergent and then to transfer it, whether before or after purification, to the polymer (Fig 3a.). This is achieved by forming first a tertiary complex, which happens spontaneously in solutions containing both types of surfactants (see refs. 12, 13, and refs. therein). The ratio of polymer to protein to be used varies depending on the molecular mass of the protein and the extension of its transmembrane surface. For small, deeply membrane inserted proteins like BR or tOmpA, a good trapping ratio is typically around 4-5 g APol per g protein (9, 14), while for a very large complex with extended extra membrane domains like cytochrome bcI , it can drop to 1.5 g per g (15). The protein will bind, ultimately, only part of the polymer added. The detergent is then removed by any convenient means (see e.g. refs. 9, 14, 16).
An alternative way of forming a MP/A8-35 complex is to start from the protein in a denaturing environment, typically urea for β-barrel MPs and SDS for α-helical ones, and to (re)fold the protein by adding the APol and removing the denaturant (17, 18; see Folding and Immobilization).
In the final, detergent-free complexes, the polymer associates specifically with the transmembrane surface of the protein (14, 19) (Fig 3b.), where it forms a compact, relatively thin layer, 1.5-2-nm thick (9). For various technical reasons, measuring precisely the amount of protein-bound APol is difficult, particularly for small proteins (see refs. 9, 12). Estimates vary from ~2 g per g protein for a small, mainly transmembrane protein like BR (9) to ~0.13 g per g for a large complex like cytochrome bcI (unpublished data). In most experiments, such as SAXS, SANS, or AUC ones, MP/A8-35 complexes behave like globular proteins (9). Upon size exclusion chromatography, however, they seem to migrate as though they were somewhat bigger than they actually are (see refs. 9, 14).