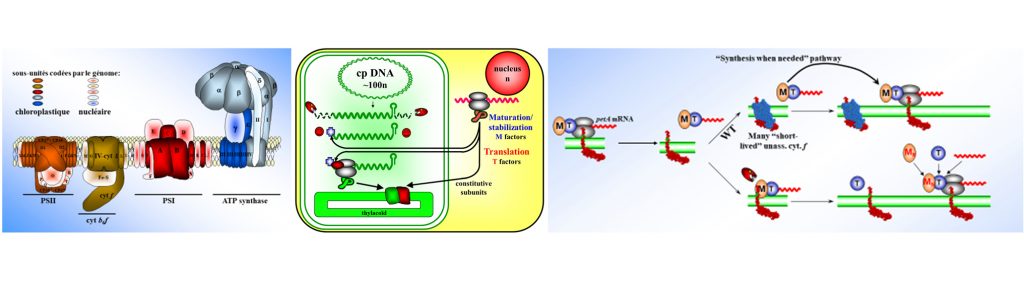
 Chloroplasts evolved from free-living cyanobacteria engulfed by a primitive eukaryotic cell about 1.5 My ago. Since then, the genes of the ancestral cyanobacterium have been either lost or transferred to the nucleus of the host cell and the chloroplast genome only encodes a fraction (50-150) of the proteins required for its own expression or for the photosynthetic function. The biogenesis of the photosynthetic apparatus and its adaptation to environmental conditions thus rely on a close cooperation of two genetic compartments. Chloroplast gene expression now relies on a mosaic of mechanisms inherited from the cyanobacterial ancestor or acquired after -endosymbiosis, to allow the host cell to tightly control the activity of its symbiont.
Chloroplasts evolved from free-living cyanobacteria engulfed by a primitive eukaryotic cell about 1.5 My ago. Since then, the genes of the ancestral cyanobacterium have been either lost or transferred to the nucleus of the host cell and the chloroplast genome only encodes a fraction (50-150) of the proteins required for its own expression or for the photosynthetic function. The biogenesis of the photosynthetic apparatus and its adaptation to environmental conditions thus rely on a close cooperation of two genetic compartments. Chloroplast gene expression now relies on a mosaic of mechanisms inherited from the cyanobacterial ancestor or acquired after -endosymbiosis, to allow the host cell to tightly control the activity of its symbiont.
We aim at a characterisation of these mechanisms and at understanding:
1) The metabolism of chloroplast RNAs by identifying its main players.
2) The nuclear control of chloroplast gene expression and how trans-acting factors recognize their chloroplast target.
3) The generality of these mechanisms, established the green lineage, in other photosynthetic clades, among which diatoms.
Selected references :
- Viola S, Cavaiuolo M, Drapier D, Eberhard S, Vallon O, Wollman FA, Choquet Y (2019) MDA1, a nucleus-encoded factor involved in the stabilization and processing of the atpA transcript in the chloroplast of Chlamydomonas. Plant J. 98:1033-1047. doi: 10.1111/tpj.14300.
- Cavaiuolo M, Kuras R, Wollman FA, Choquet Y, Vallon O (2017) Small RNA Profiling in Chlamydomonas: Insights into Chloroplast RNA Metabolism. Nucl. Ac. Res., 45:10783-10799. doi: 10.1093/nar/gkx668.
- Boulouis A, Drapier D, Razafimanantsoa H, Wostrikoff K, Tourasse NJ, Pascal K, Girard-Bascou J, Vallon O, Wollman F-A, Choquet Y (2015) Spontaneous dominant mutations in Chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell, 27: 984-1001.
